RandomSampleImputer#
The RandomSampleImputer() replaces missing data with a random sample extracted from the
variable. It works with both numerical and categorical variables. A list of variables
can be indicated, or the imputer will automatically select all variables in the train
set.
Note
The random samples used to replace missing values may vary from execution to execution. This may affect the results of your work. Thus, it is advisable to set a seed.
Setting the seed#
There are 2 ways in which the seed can be set in the RandomSampleImputer():
If seed = 'general' then the random_state can be either None or an integer.
The random_state then provides the seed to use in the imputation. All observations will
be imputed in one go with a single seed. This is equivalent to
pandas.sample(n, random_state=seed) where n is the number of observations with
missing data and seed is the number you entered in the random_state.
If seed = 'observation', then the random_state should be a variable name
or a list of variable names. The seed will be calculated observation per
observation, either by adding or multiplying the values of the variables
indicated in the random_state. Then, a value will be extracted from the train set
using that seed and used to replace the NAN in that particular observation. This is the
equivalent of pandas.sample(1, random_state=var1+var2) if the seeding_method is
set to add or pandas.sample(1, random_state=var1*var2) if the seeding_method
is set to multiply.
For example, if the observation shows variables color: np.nan, height: 152, weight:52, and we set the imputer as:
RandomSampleImputer(random_state=['height', 'weight'],
seed='observation',
seeding_method='add'))
the np.nan in the variable colour will be replaced using pandas sample as follows:
observation.sample(1, random_state=int(152+52))
For more details on why this functionality is important refer to the course Feature Engineering for Machine Learning.
You can also find more details about this imputation in the following notebook.
Note, if the variables indicated in the random_state list are not numerical
the imputer will return an error. In addition, the variables indicated as seed
should not contain missing values themselves.
Important for GDPR#
This estimator stores a copy of the training set when the fit() method is
called. Therefore, the object can become quite heavy. Also, it may not be GDPR
compliant if your training data set contains Personal Information. Please check
if this behaviour is allowed within your organisation.
Below a code example using the House Prices Dataset (more details about the dataset here).
First, let’s load the data and separate it into train and test:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from feature_engine.imputation import RandomSampleImputer
# Load dataset
data = pd.read_csv('houseprice.csv')
# Separate into train and test sets
X_train, X_test, y_train, y_test = train_test_split(
data.drop(['Id', 'SalePrice'], axis=1),
data['SalePrice'],
test_size=0.3,
random_state=0
)
In this example, we sample values at random, observation per observation, using as seed the value of the variable ‘MSSubClass’ plus the value of the variable ‘YrSold’. Note that this value might be different for each observation.
The RandomSampleImputer() will impute all variables in the data, as we left the
default value of the parameter variables to None.
# set up the imputer
imputer = RandomSampleImputer(
random_state=['MSSubClass', 'YrSold'],
seed='observation',
seeding_method='add'
)
# fit the imputer
imputer.fit(X_train)
With fit() the imputer stored a copy of the X_train. And with transform, it will extract
values at random from this X_train to replace NA in the datasets indicated in the transform()
methods.
# transform the data
train_t = imputer.transform(X_train)
test_t = imputer.transform(X_test)
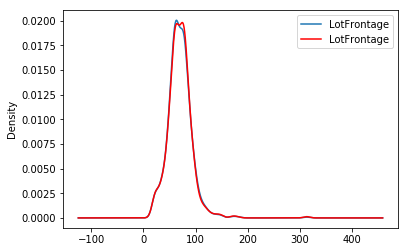
The beauty of the random sampler is that it preserves the original variable distribution:
fig = plt.figure()
ax = fig.add_subplot(111)
X_train['LotFrontage'].plot(kind='kde', ax=ax)
train_t['LotFrontage'].plot(kind='kde', ax=ax, color='red')
lines, labels = ax.get_legend_handles_labels()
ax.legend(lines, labels, loc='best')
Additional resources#
In the following Jupyter notebook you will find more details on the functionality of the
RandomSampleImputer(), including how to set the different types of seeds.
All Feature-engine notebooks can be found in a dedicated repository.
And finally, there is also a lot of information about this and other imputation techniques in this online course:

Feature Engineering for Machine Learning#
Or read our book:

Python Feature Engineering Cookbook#
Both our book and course are suitable for beginners and more advanced data scientists alike. By purchasing them you are supporting Sole, the main developer of Feature-engine.
